Charting a path to better cell models of the intestine
Researchers generate an intestinal cell mimic that can be harnessed in studies of diseases such as inflammatory bowel disease.
For many years, drug development has relied on simplified and scalable cell culture models to find and test new drugs for a wide variety of diseases. However, cells grown in a dish are often a feint representation of healthy and diseased cell types in vivo. This limitation has serious consequences: Many potential medicines that originally appear promising in cell cultures often fail to work when tested in patients, and targets may be completely missed if they do not appear in a dish.
A highly collaborative team of researchers from the Harvard-MIT Program in Health Sciences and Technology (HST) and Institute for Medical Engineering and Science (IMES) at MIT recently set out to tackle this issue as it relates to a type of cell found in the intestine that is implicated in inflammatory bowel disease (IBD). In new work, the team was able to generate an intestinal cell that is a substantially better mimic of the real cell and can therefore be used in studies of diseases such as IBD. They reported their findings in a recent issue of BMC Biology.
The team was led by Ben Mead, a doctoral student in the HST Medical Engineering and Medical Physics Program; Jeffrey Karp, a professor at Brigham and Women’s Hospital, working closely with Jose Ordovas-Montanes, a postdoc in the lab of Pfizer-Laubach Career Development Assistant Professor Alex K. Shalek in the MIT Department of Chemistry; and the labs of MIT professor of biological engineering Jim Collins, Institute Professor Robert Langer, and scientists from the Broad Institute of Harvard and MIT and Koch Institute for Integrative Cancer Research.
Understanding genetic risk at the level of single cells
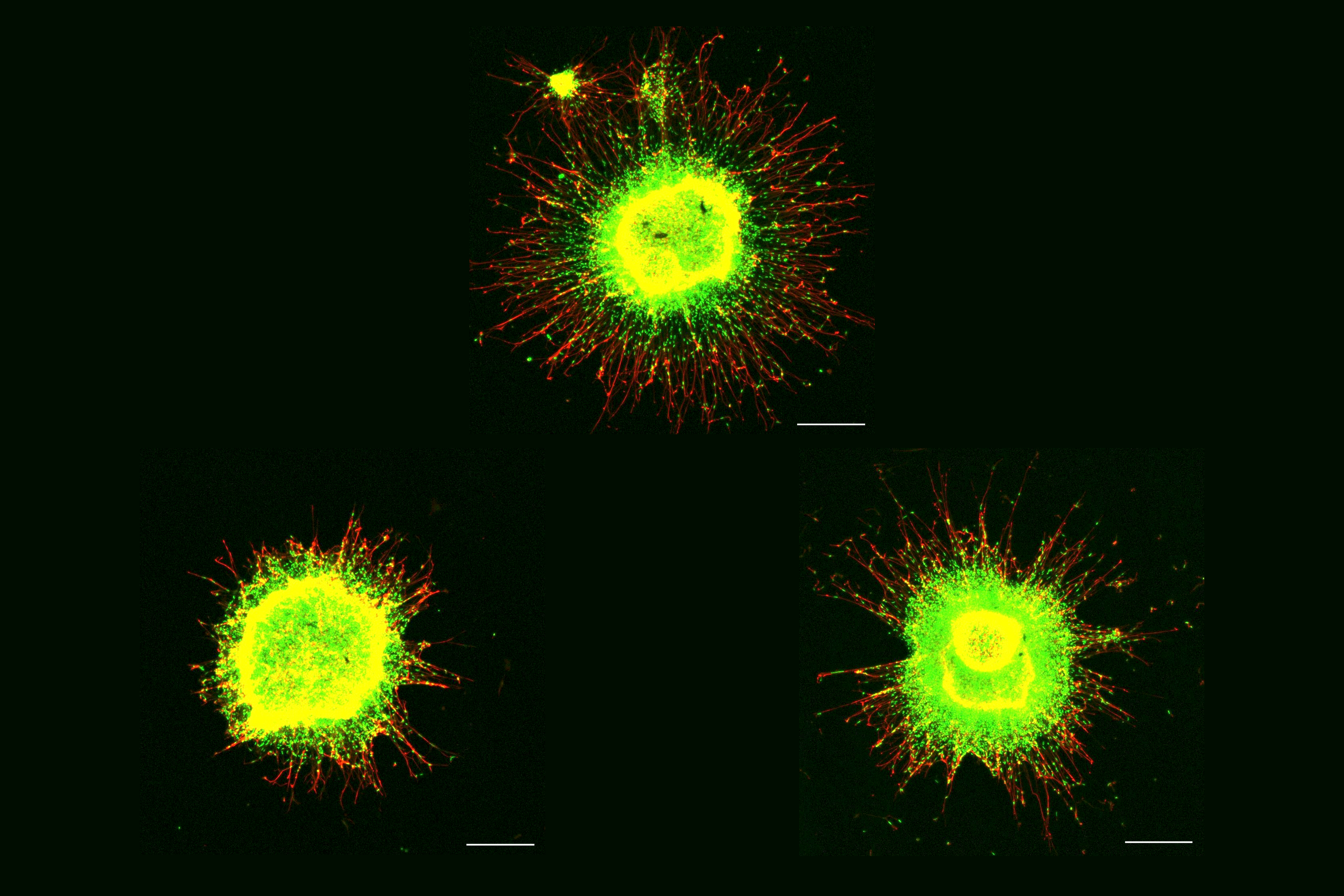
This study was catalyzed by the new technology of high-throughput single-cell RNA-sequencing, which enables transcriptome-wide profiling of tissues at the level of individual cells. Through the lens of single-cell RNA-sequencing, scientists are now able to ‘map’ our single cells and potentially the changes which give rise to disease. The team of researchers turned this method towards determining how well an existing cell culture model mimics a particular type of cell within the body, comparing two single cell ‘maps’: one of a mouse’s small intestine, and another of an adult stem cell-derived model of the small intestine, known as an organoid.
They used these maps to isolate a single cell type and ask how well the organoid-derived cell matched its natural counterpart. “Based on the differences between model and actual cell, we utilized a computationally driven bioengineering approach to improve the fidelity of the model.” said Karp. “We believe this approach may be key to unlocking the next generation of therapeutic development from cellular models, including those made from patient-derived stem cells.”
Individual genes can alter one’s risk of developing diseases such as Crohn’s disease, a type of IBD. One active area of research is understanding where these genes act in a tissue in order to further our understanding of disease mechanisms and propose novel therapeutic interventions. To address this, techniques are needed to reliably map “risk” genes not only within an affected tissue, but to individual cells, to properly surmise if a drug screen can correct a faulty gene or potentially improve a patient’s condition.
Single-cell RNA-sequencing at scale, a revolutionary technique pioneered for low-input clinical biopsies at MIT between Alex K. Shalek’s and Chris Love’s group, now allows researchers to deconstruct a tissue into its elemental components — cells — and identify the key patterns of gene expression which specify each cell type. The ability to efficiently profile tens of thousands of cells economically has unlocked the possibility to identify critical cell types in tissues whose genetic makeup had previously been difficult to discern.
Using single-cell “maps” to re-orient the development of a key cell type
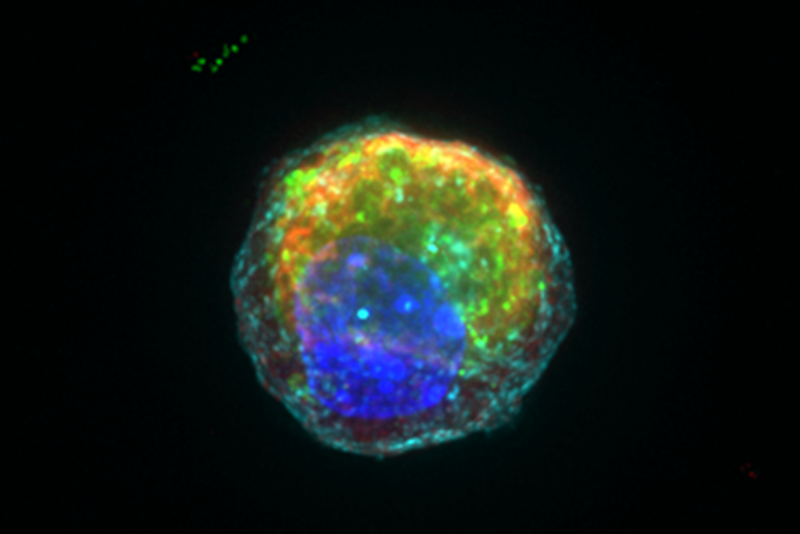
Mapping tissues, such as the small intestine, is highly important in understanding where specific “risk” genes are acting. However, the key advances required to translate findings to the clinic will inevitably be through representative models for the cell types identified as interpreting genes and displaying a disease phenotype. One key IBD-relevant cell type already implicated through genetic studies is known as the Paneth cell, responsible for a key anti-microbial role in the small intestine and defending the stem cell niche.
When adult intestinal stem cells are grown in a dish, they self-organize into remarkable structures known as intestinal organoids: 3-D cellular structures that contain many of the cell types found in a real intestine. Nevertheless, how these intestinal organoids correspond to the bona fide cell types found in the intestine has proven challenging for researchers to tackle. To directly address this question, Shalek suggested a “quick” experiment to Mead, which then gave rise to the fruitful collaboration between the labs.
Mead and Ordovas-Montanes developed a single-cell map of the true characteristics of small intestinal cell types as found within the mouse and, when comparing them to what a map of the intestinal-derived organoid looks like, identified several differences, particularly within the key IBD-relevant cell type known as the Paneth cell. Since the field’s map of an organoid didn’t quite correspond to the real tissue, it may have led them astray in the hunt for drug targets.
Fortunately, through their single-cell data, the team was able to learn how the maps were mis-aligned, and “correct” the developmental pathways which were missing in the dish. As a result, they were able to generate a Paneth cell that is a substantially better mimic of the real cell and can now function to kill bacteria and support the neighboring stem cells which give rise to them.
Translational opportunities afforded by improved representations of tissues
“With this improved cell in-hand, we are now developing a screening platform that will allow us to target relevant Paneth cell biology,” says Mead, who plans to continue the work he started as a postdoc in Shalek’s group.
Their approach for generating physiologically faithful intestinal cell types is a major technological advance that will provide other researchers a powerful tool to further their understanding of the specialized cell states of the epithelial barrier. “As we begin to understand which cell types specifically express genes that alter risk for IBD, it will be critical to ensure the disease models provide an accurate representation of that cell type,” says Ordovas-Montanes.
“We want to make better cell models to not only understand basic disease biology, but also to fast-track development of therapeutics” says Mead. “This research will have impact beyond the intestinal organoid community as organoids are increasingly employed for liver, kidney, lung, and even brain research, and our approach can be generalized for relating and aligning the cell types found in vivo with the models generated from these tissues.”